

It takes very long time to run a single apsimx file with lots of simulations (e.g. wheat.apsimx in the release contains more than 4000 simulations and takes more than 20 min to run all simulations). Multiple experiments are used to test a crop model and organise into an apsimx file. SuppressPackageStartupMessages(library(tidyverse)) SuppressPackageStartupMessages(library(rapsimng)) Calibrate and validate the leaf appearance process through comparing simulated and observed values.Run tasks using cluster to obtain simulated values for factorial simulations.Generate parameter spaces and prepare task list for parallel calculation.Split apsimx into individual simulations for each treatment except cultivar factor.The method of factorial simulations can be separated into multiple steps including The codes in this page might not be reproducible.
The method heavily depends on the available CPU resources and infrastructure of cluster which might not be suitable for every situation. All 77 cultivars are simultaneously calibrated and validated in this process. In this page, I presented a method using factorial simulations to calibrate and validate the leaf appearance process for wheat validation set in the APSIM Next Generation. However, it is time consuming to calibrate lots of cultivars using observations from multiple experiments.
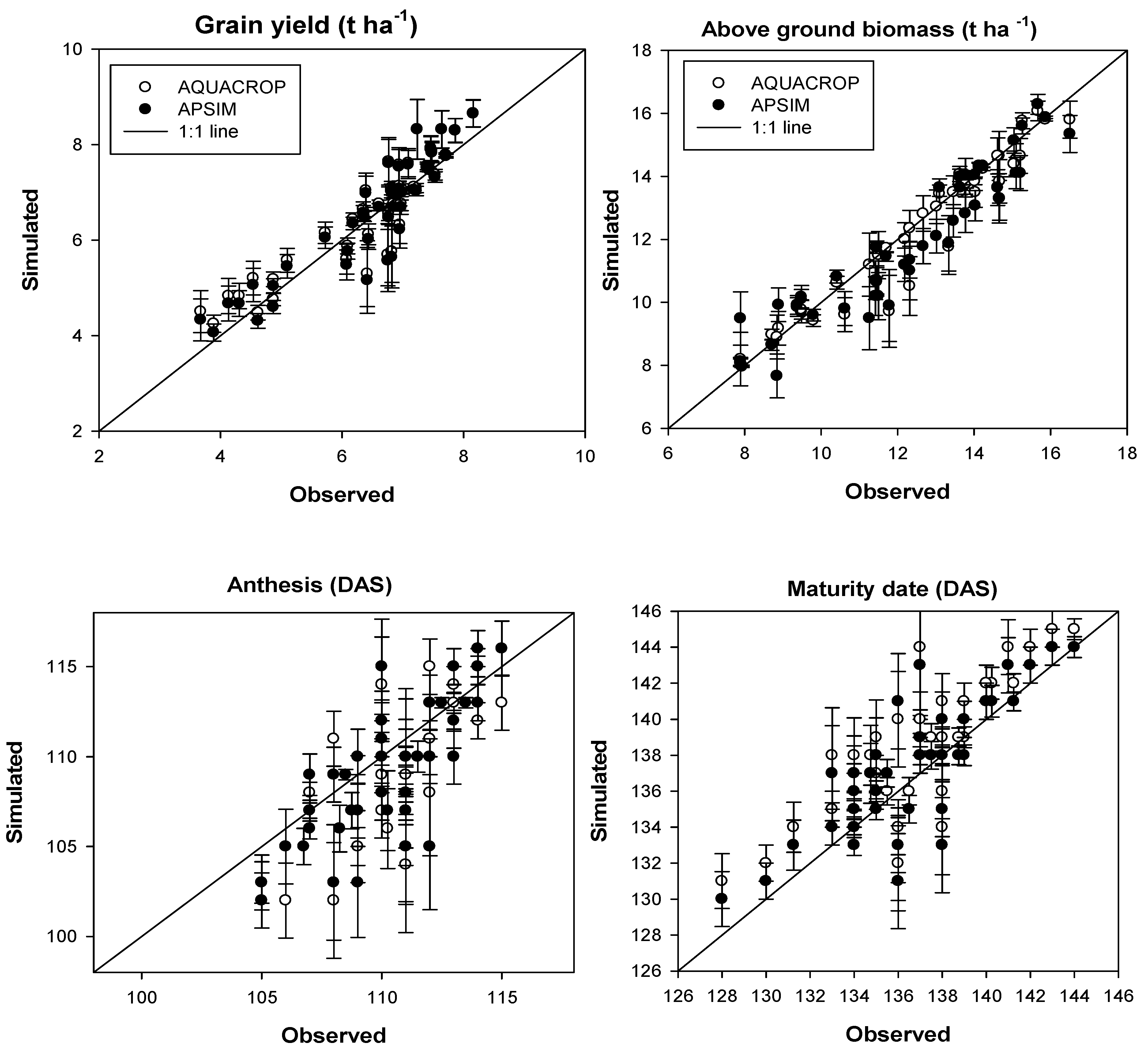
Optimization of parameter values in the crop model is the key step to utilise models to simulate the field experiments.


 0 kommentar(er)
0 kommentar(er)
